https://blog.tensorflow.org/2019/03/intelligent-scanning-using-deep-learning.html?hl=ur
https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEhyNOcGmfZqpX2hZ4wGV1wTneR3YIo4eqQybcJ3DfdKk7FdW3fMQmc2ZO-oXEt8_uvfewDBf1N8vMkWVjB3Ya_DAZiqoSz2oyY7ZSipFBARPzM-sLX-TBo7C_S6le6zt6IA-GEB1vXSzWQ/s1600/figure1.gif
Posted by Jason A. Polzin, PhD GM Applications and Workflow, GE Healthcare Global Magnetic Resonance Imaging
Here we describe our experience using TensorFlow to train a neural network to identify specific anatomy during a brain magnetic resonance imaging (MRI) exam to help improve speed and consistency.
Introduction
MRI (Figure 1.) is a 3D imaging technique that allows clinicians to visualize structures in the body non-invasively and without ionizing radiation. MRI is a widely used and powerful imaging modality due to its superior contrast between “soft” tissues, e.g. gray matter, white matter, and CSF, as well as its unique ability to not only visualize anatomical structures but also depict physiology and function, e.g. blood flow, perfusion, and diffusion.
One of the key strengths of MRI is being able to image specific locations in the body at an orientation best suited for the purpose of the exam. This means that the operator must plan these scans carefully to yield the best possible images uniquely oriented for each patient to visualize the specific structures that may be of interest.
 |
| Figure 1. The Power of MRI |
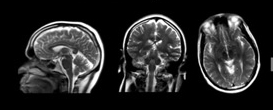 |
| Figure 2. Example of a localizer acquisition (3 Plane) |
The Challenge
As part of the MRI exam, the scan operator first acquires a set of low-resolution “localizer” images (Figure 2) from which approximate location and orientation of desired landmarks can be identified. These anatomical references are then used to manually plan the exact locations, orientation, and required coverage for images that will be used for the high-resolution scans that are used for diagnosis. This procedure is complicated as the orientation, location, and coverage needs to be correct in all three spatial dimensions.
The quality and consistency of positioning and orientation of the slices relies heavily on the skill and experience of the scan operator. This process can be time-consuming and difficult, especially for complex anatomies. As a result, there can be inconsistencies from scan operator to scan operator. This lack of consistency can make the job of the radiologist in interpreting these images more difficult especially when a patient is being scanned as a follow up to previous MRI exam and they are trying to identify subtle changes in anatomy or disease progression over time. In the worst-case scenario, the correct images are not obtained, and the patient must return for another procedure.
The Solution
To aid the scan operator we developed a deep-learning (DL) based framework for intelligent MRI slice placement (ISP) for several commonly used brain landmarks. Our approach determines plane orientations automatically using only the standard clinical localizer images. This allows the scan operator to consistently get patient-specific slice orientations for multiple anatomical brain landmarks: anterior commissure-posterior commissure, orbitomeatal, entire visual pathway including multiple orientations through the orbits and optic nerve, pituitary, internal auditory canal, hippocampus, and circle of Willis (angiography).
We utilized the TensorFlow library with the
Keras interface to implement the DL based framework for ISP. We choose TensorFlow since it provided ready support for 2D and 3D Convolutional Neural Networks (CNN), which is the primary requirement for medical image volume processing, and the Keras API made it easy to rapidly develop and test our ideas.
Unlike prior approaches to automate slice placement, our ISP framework uses deep-learning (DL) to determine the necessary plane(s) without the need for explicit delineation of landmark structures or customization of the localizer images. It can also warn the user in case the localizer images do not have enough information for automatically determining best scan planes.
The localizer images are very low-resolution images with limited brain coverage and several fine structures are not easily identified. An approach to directly determine the orientation of structures of interest would be very hard to implement using classical image analysis as both identification and generation of features to achieve this would be very error prone. Moreover, as compared to the classical approaches, a DL-based approach is less affected by factors that affect MRI image quality or appearance. This makes our DL approach robust to differences in MRI hardware, site specific parameter settings, and in general patient positioning across different clinics and hospitals.
Another key advantage of our generalized DL-based approach is that it can be easily extended across other anatomies. Using the exact same architecture, but new training data, we can develop automated techniques for new anatomies, e.g. knee, spine, etc., which drastically reduces development time.
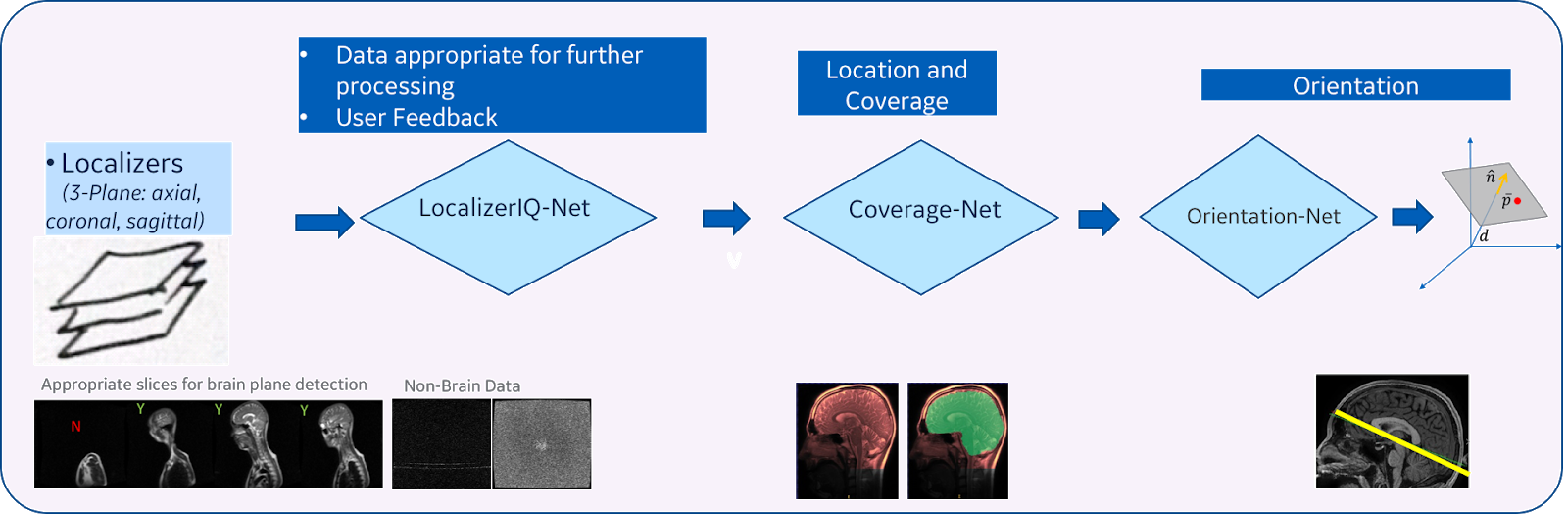 |
| Figure 3. DL-based intelligent scan placement framework |
Our method consists of three main steps (Figure 3):
- Image quality inspection: In the first step, we check if the given localizer image is suitable to identify the plane for the desired anatomy. This is achieved by using a fiver layer, dyadic reduction regular CNN classification network model (that we call “LocalizerIQ-Net”) to identify slices with relevant brain-anatomy, slices with artifacts and irrelevant slices. If the localizer image is not suitable for ISP, relevant feedback is provided to the scan operator. We used the built-in TensorFlow functions for image manipulation to achieve data augmentation during the training of LocalizerIQ-Net.
- Identification of anatomy coverage: Next, we locate the spatial-extent of the desired anatomy (brain) in the localizer images by incorporating a shape-based semantic image segmentation U-Net DL model (called “Coverage-Net”). This helps the next processing steps to be robust to changes in imaging parameter settings across hospitals and clinics as well as changes in shapes and sizes of the patient’s anatomy.
- Identification of precise plane orientation and location: For each desired anatomic structure (example, optic nerve), we find the scan-plane that is best suited to image that structure using one or more image segmentation 3D U-Net models (called “Orientation-Net”). Orientation-Net directly segments the desired-planes on localizer images, which is then used to compute the orientation and location.
The entire process takes ~3.5 seconds on a high-performance CPU. The training and testing data for these DL models came from more than 1300 subjects and were acquired using various GE scanner models and field strengths from several clinical sites across the globe. The imaging data used for training had variations in contrast, image resolution and other scan settings across the patients. Examples of ground truth labels are shown in Figure 4.
 |
| Figure 4. Examples of GT labels for coverage and orientation for brain IPS. (a), (b): Coverage mask for sagittal and coronal orientation; (c), (d): Mid-sagittal plane mask (red) with the fitted plane shown as a white line. (e): AC and PC labels (arrows) along AC-PC plane line (white line). (f): Optic Nerve label (red label) along with optic nerve plane. (g): Hippocampus label along with hippocampus plane. |
LocalizerIQ-Net was trained using a total of 29,000 images (with image augmentation) and tested using 700 images. LocalizerIQ-Net obtained a classification accuracy of 99.2% on the independent test images. Coverage-Net and Orientation-Net were trained with a total of 21,770 volumes (with augmentation) and tested using 505 volumes. Orientation-Net obtained mean distance error of < 1 mm and angle error < 3° for desired structures, which was considered as acceptable for clinical use by radiologists.
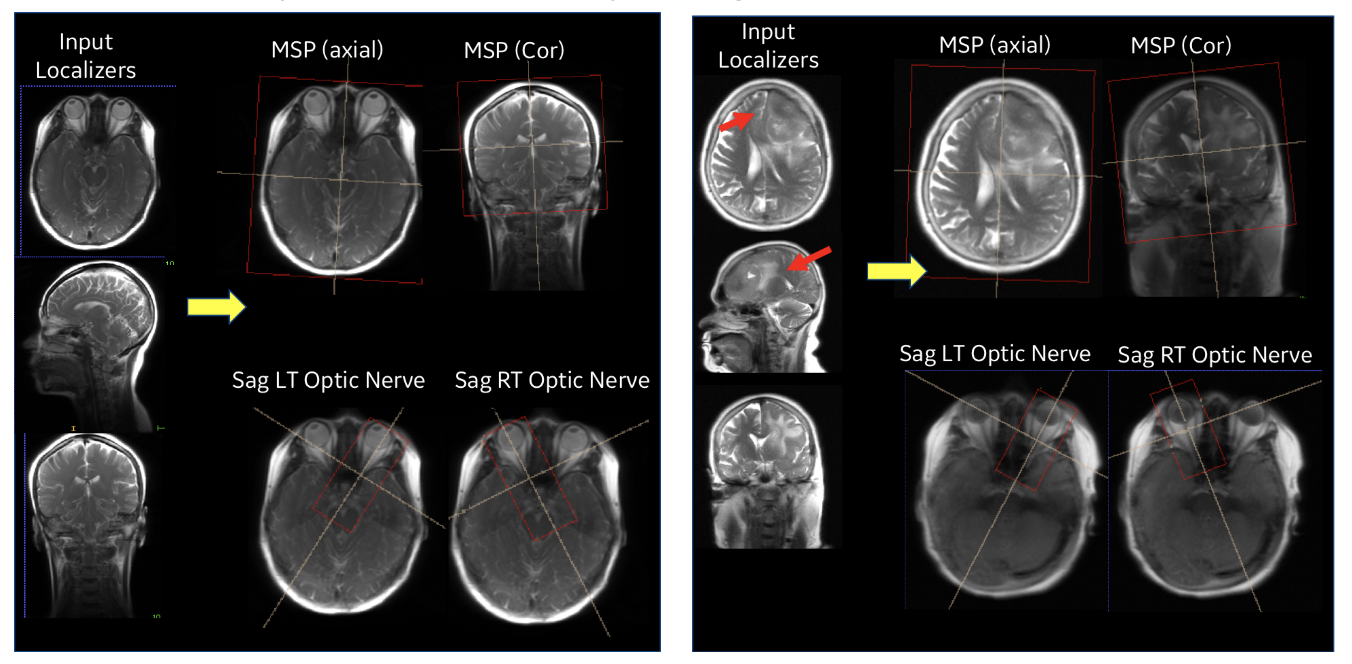 |
| Figure 5. Performance of ISP in a normal subject case (left) and with pathology (right). |
Our entire ISP workflow is implemented and integrated on various GE scanners and has been evaluated at several clinical sites. ISP provided accurate slice placement for various landmarks planes on localizer images (Figure 5 (left)). We have shown that our method works even when there is significant pathology in the image (Fig. 5 (right)). Figure 6 demonstrates the utility of our method where the patient’s head is in three very different positions in three exams (as shown in the top row), but the acquired image (bottom row) is consistent across the three different head poses. This is critical for longitudinal exams where data consistency in terms of landmark location helps improve radiologist image reading efficiency.
 |
| Figure 6. Consistent brain image orientation irrespective of various head rotations. |
Benefits of TensorFlow
We chose TensorFlow as our development and deployment platform for the following reasons:
- Support for 2D and 3D Cascaded Neural Networks (CNN) which is the primary requirement for medical image volume processing
- Extensive built-in library functions for image manipulation and optimized tensor computations
- Extensive open-source user and developer community which supported latest algorithm implementations and made them readily available
- Continuous development with backward compatibility making it easier for code development and maintenance
- Stability of graph computations made it attractive for product deployment
- Keras interface was available which significantly reduced the development time: This helped in generating and evaluating different models based on hyper-parameter tuning and determining the most accurate model for deployment.
- Deployment was done using a TensorFlow Serving CPU based docker container and RestAPI calls to process the localizer once it is acquired.
AIRx™
The GE Healthcare product that incorporates the Deep Learning based Intelligent Slice Placement, is called
AIRx or Artificial Intelligence Prescription. Here’s a
video demonstrating the capability.
Our tests have shown that with
AIRx, the time it takes for the scan operator to determine the locations and orientations for the desired scan planes for a brain MRI can be reduced by 40% to 60%. Furthermore, we have also seen a reduction in errors and improved accuracy. This could result in shorter overall exams, with reduced chance of patient call back, and improved exam diagnostic quality.
What's Next
We have started work using the same TensorFlow based platform to automate determination of the slice orientations for other common MR procedures including the knee and spine. The capabilities enabled by DL using the TensorFlow platform will also allow us to automate other manual tasks that are part of the overall MRI procedure as part of our goal to build a fully autonomous MRI scanner. This will allow the scan operator to focus on the patient rather than the specific machine settings required to get the best image quality for each patient.
Acknowledgments
A big thanks to the team from GE Healthcare and GE Global Research Center who are responsible for developing this feature and specifically Chitresh Bhushan, Thomas Foo, Dawei Gui, Dattesh Shanbhag, and Sheila Washburn who all contributed to this blog post.







